Credit: Nicola Ferrari/Getty Images
- Specificity
Each antibody is attached with an individual DNA barcode.
Add all your antibodies at once.
- Resolution
The patented Tag-DNA amplification mechanism provide single cell resolution
- Biomarkers
Detect ~1000 biomarkers without cycle
- Specific Instruments Free
Based on spatial transcriptomics platforms such as Stereo-seq from Complete Genomics.
Sequencing-based Spatial Proteomics
with Tag-DNA Amplification
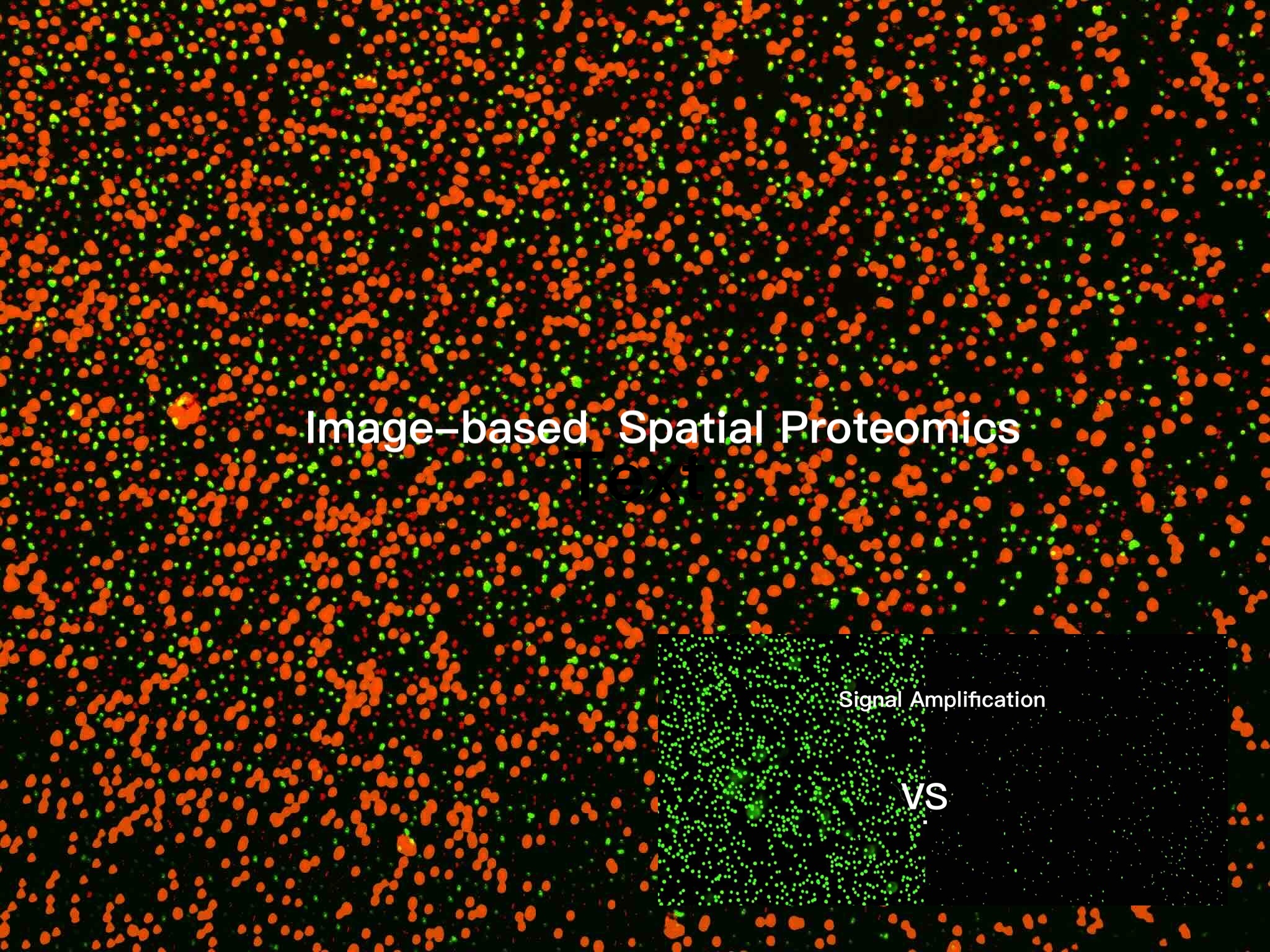
Image-based Spatial Proteomics
with Signal-DNA Amplification
- Specificity
Each antibody is attached with an individual DNA barcode.
Add all your antibodies at once.
- Sensitivity
The patented signal amplification mechanism provide 30x greater signal-to-noise ratios than conventional IF
- Biomarkers
Detect ~100 biomarkers without cross-reactivity.
- Specific Instruments
Use Your Existing Fluorescent Imaging Instrumentation
Spatial proteomics is an umbrella term that covers a broad swath of immunohistochemistry-based methods including, but not limited to, cyclic immuno- fluorescence (cycIF), co-detection by indexing (CODEX), iterative bleaching extends multi- plexity (IBEX), multiplexed ion beam imaging (MIBI) and imaging mass cytometry (IMC).
These approaches can be used to generate highly multiplexed images of specimens such as tissue and organ slices to understand their protein composition and spatial organization and are the basis of many global atlas projects.
Why oligonucleotide based cycIF?
In each cycle, the removal of the complementary oligos containing the fluorophores is very simple, enabling the next round of imaging.
Why oligonuleotide amplification?
Oligonucleotide amplification results in the extension of the DNA barcode.
Therefore, more fluorescent probes are bound, resulting in a higher signal-to-noise ratio.
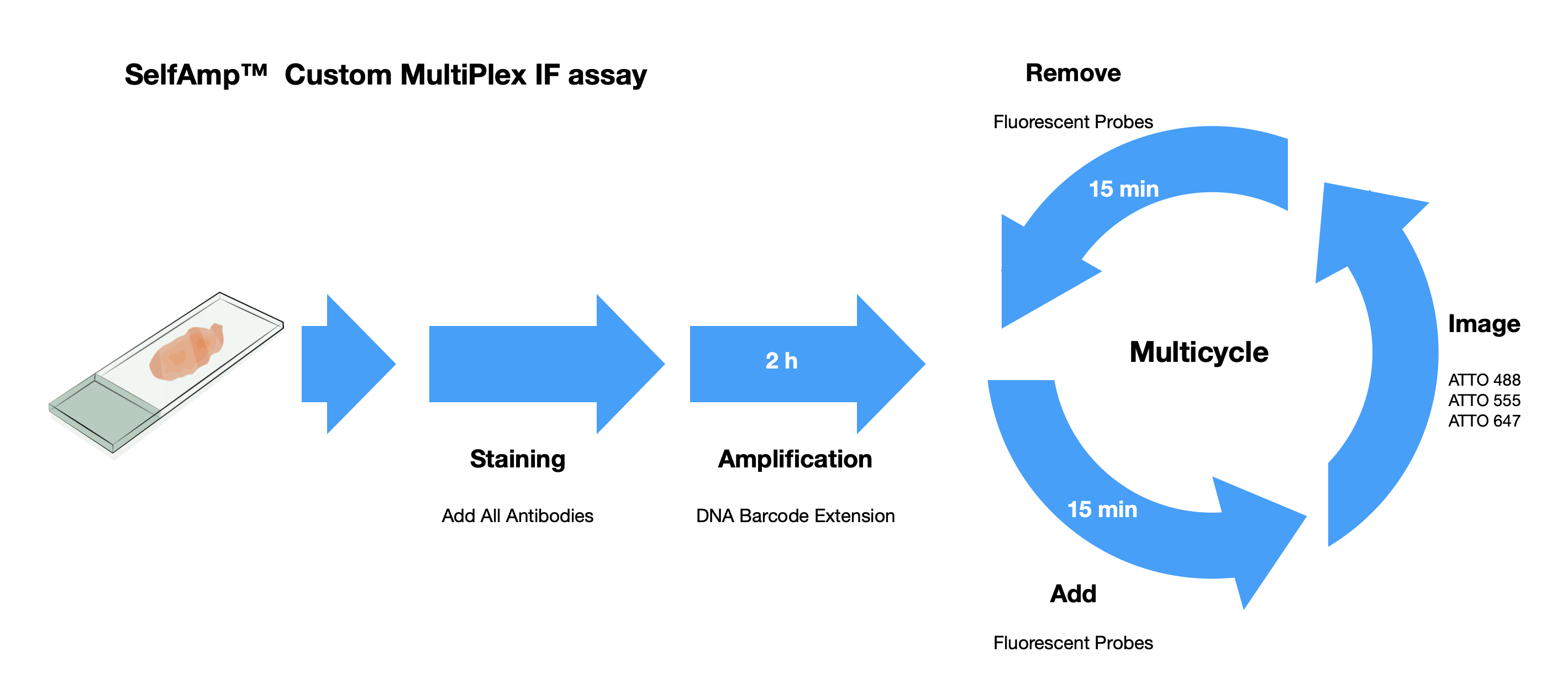
Why SelfAmp technology ?
SelfAmp™ technology utilizes a mixture of DNA barcoded antibodies. After binding to the antigens and amplification, those antibodies are attached by fluorescent probes, which can be visualized using a fluorescence microscope.
Due to the spontaneous amplification without the need for any additional nucleic acid strands, there is no Cross-Reactivity within the barcoded DNA. So we can mix as many DNA barcode antibodies as possible.
With our validated technology, we eliminate the limitation on the number of protein targets that can be detected in multiplex immunofluorescence experiments.
Copyright © 2024 Haoqinbio,Inc. All Rights Reserved